Note
Go to the end to download the full example code.
Equilibrating an MD box of acetonitrile#
Goals#
In this tutorial we learn how to perform a thermal equilibration of a box of acetonitrile molecules using ASE. We will:
build a coarse-grained CH₃–C≡N triatomic model.
set up a periodic box at the experimental density (298 K).
apply rigid-molecule constraints.
use the ACN force field.
equilibrate with Langevin dynamics.
scale small box of 27 molecules to 216 molecules.
ACN model#
The acetonitrile force field implemented in ASE (ase.calculators.acn)
is an interaction potential between three-site linear molecules. The atoms
of the methyl group are treated as a single site centered on the methyl
carbon (hydrogens are not explicit). Therefore:
assign the methyl mass to the outer carbon (
m_me),use the atomic sequence Me–C–N repeated for all molecules,
keep molecules rigid during MD with
FixLinearTriatomic.
import matplotlib.pyplot as plt
import numpy as np
import ase.units as units
from ase import Atoms
from ase.calculators.acn import ACN, m_me, r_cn, r_mec
from ase.constraints import FixLinearTriatomic
from ase.io import Trajectory
from ase.md import Langevin
from ase.visualize.plot import plot_atoms
Step 1: molecule#
Build one CH3–C≡N as a linear triatomic “C–C–N”. The first carbon is the methyl site; we assign it the CH3 mass. Rotate slightly to avoid perfect alignment with the cell axes.
pos = [[0, 0, -r_mec], [0, 0, 0], [0, 0, r_cn]]
atoms = Atoms('CCN', positions=pos)
atoms.rotate(30, 'x')
masses = atoms.get_masses()
masses[0] = m_me
atoms.set_masses(masses)
mol = atoms.copy()
mol.set_pbc(False)
mol.set_cell([20, 20, 20])
mol.center()
fig, ax = plt.subplots(figsize=(4, 4))
plot_atoms(
mol,
ax=ax,
rotation='30x,30y,0z',
show_unit_cell=0,
radii=0.75,
)
ax.set_axis_off()
plt.tight_layout()
plt.show()
Step 2: Set up small box of 27-molecules#
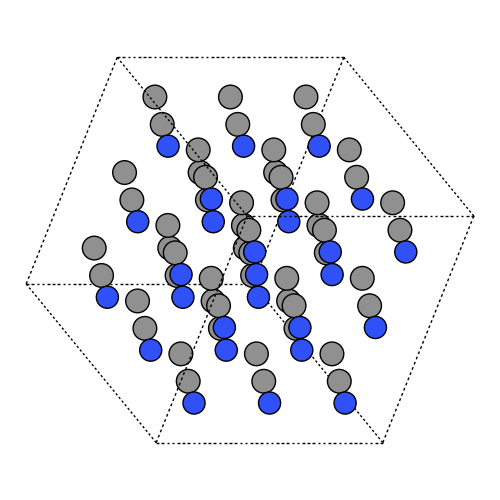
Match density 0.776 g/cm^3 at 298 K. Compute cubic box length L from mass and density. Build 3×3×3 supercell and enable PBC.
density = 0.776 / 1e24 # g / Å^3
L = ((masses.sum() / units.mol) / density) ** (1 / 3.0)
atoms.set_cell((L, L, L))
atoms.center()
atoms = atoms.repeat((3, 3, 3))
atoms.set_pbc(True)
box27 = atoms.copy()
fig, ax = plt.subplots(figsize=(5, 5))
plot_atoms(
box27,
ax=ax,
rotation='35x,35y,0z',
show_unit_cell=2,
radii=0.75,
)
ax.set_axis_off()
plt.tight_layout()
plt.show()
Step 3: Set constraints#
Keep each “C–C–N” rigid during MD using FixLinearTriatomic.
Step 4: MD run for 27-molecules system#
Assign ACN with cutoff = half the smallest box edge. Langevin MD at 300 K, 1 fs timestep. Save a frame every step.
True
Step 5: scale system size to 216 molecules#
Repeat 2×2×2 to reach 216 molecules. Reapply constraints and update the ACN cutoff for the new cell.
Step 6: MD run for 216-molecules system#
tag = 'acn_216mol_300K'
md = Langevin(
atoms,
2 * units.fs,
temperature_K=300,
friction=0.01,
logfile=tag + '.log',
)
traj = Trajectory(tag + '.traj', 'w', atoms)
md.attach(traj.write, interval=10)
times_ps, epots, ekins, etots, temps = [], [], [], [], []
sample_interval = 10 # sample every 10 MD steps for lighter plots
def sample():
# Time in ps (same as MDLogger: dyn.get_time() / (1000 * units.fs))
t_ps = md.get_time() / (1000.0 * units.fs)
ep = atoms.get_potential_energy() # eV total
ek = atoms.get_kinetic_energy() # eV total
T = atoms.get_temperature() # K
times_ps.append(t_ps)
epots.append(ep)
ekins.append(ek)
etots.append(ep + ek)
temps.append(T)
# initial sample at t=0
sample()
md.attach(sample, interval=sample_interval)
md.run(1000) # 6 ps @ 2 fs
True
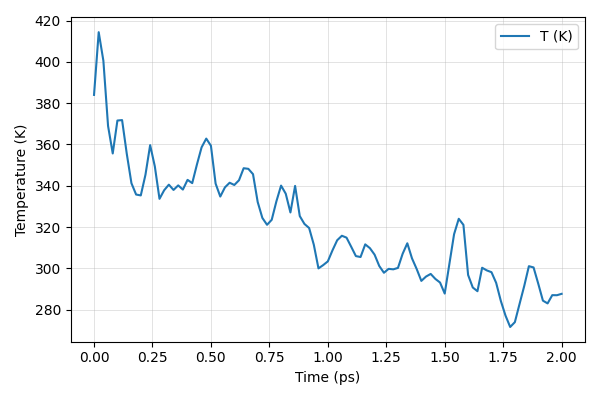
Plot Instantaneous temperature vs time. Does the system equilibrated well? What is the average temperature? Should we run longer simulations?
Next steps#
View trajectories:
Plot other thermodynamic quantities (p.e., k.e., and total energy).
Total running time of the script: (4 minutes 7.694 seconds)
