Note
Go to the end to download the full example code.
Calculating Delta-values#
In this tutorial we compare the equation-of-state (EOS) calculated for 7 FCC
metals using values from EMT, WIEN2k and
experiment. Each EOS is described by three parameters:
volume per atom
bulk modulus
pressure derivative of the bulk modulus
Differences between two EOS’s can be measured by a single \(\Delta\) value defined as:
where \(E_n(V)\) is the energy per atom as a function of volume.
The \(\Delta\) value can be calculated using the
ase.utils.deltacodesdft.delta() function:
- ase.utils.deltacodesdft.delta(v1: float, B1: float, Bp1: float, v2: float, B2: float, Bp2: float, symmetric=True) float[source]
Calculate Delta-value between two equation of states.
See also
- Parameters:
v1 (float) – Volume per atom.
v2 (float) – Volume per atom.
B1 (float) – Bulk-modulus (in eV/Ang^3).
B2 (float) – Bulk-modulus (in eV/Ang^3).
Bp1 (float) – Pressure derivative of bulk-modulus.
Bp2 (float) – Pressure derivative of bulk-modulus.
symmetric (bool) – Default is to calculate a symmetric delta.
- Returns:
delta – Delta value in eV/atom.
- Return type:
See also
Collection of ground-state elemental crystals: DeltaCodesDFT
Equation-of-state module:
ase.eos
We get the WIEN2k and experimental numbers from the DeltaCodesDFT ASE-collection and we calculate the EMT EOS using this script:
from pathlib import Path
import numpy as np
from ase.calculators.emt import EMT
from ase.collections import dcdft
from ase.io import Trajectory
trajfiles = []
for symbol in ['Al', 'Ni', 'Cu', 'Pd', 'Ag', 'Pt', 'Au']:
trajfile = Path(f'{symbol}.traj')
with Trajectory(trajfile, 'w') as traj:
for s in range(94, 108, 2):
atoms = dcdft[symbol]
atoms.set_cell(atoms.cell * (s / 100) ** (1 / 3), scale_atoms=True)
atoms.calc = EMT()
atoms.get_potential_energy()
traj.write(atoms)
trajfiles.append(trajfile)
And fit to a Birch-Murnaghan EOS:
import json
from ase.eos import EquationOfState as EOS
from ase.io import read
def fit(symbol: str) -> tuple[float, float, float, float]:
V = []
E = []
for atoms in read(f'{symbol}.traj@:'):
V.append(atoms.get_volume() / len(atoms))
E.append(atoms.get_potential_energy() / len(atoms))
eos = EOS(V, E, 'birchmurnaghan')
eos.fit(warn=False)
e0, B, Bp, v0 = eos.eos_parameters
return e0, v0, B, Bp
data = {} # Dict[str, Dict[str, float]]
for path in trajfiles:
symbol = path.stem
e0, v0, B, Bp = fit(symbol)
data[symbol] = {
'emt_energy': e0,
'emt_volume': v0,
'emt_B': B,
'emt_Bp': Bp,
}
Path('fit.json').write_text(json.dumps(data))
960
Result for Pt using EMT:
import matplotlib.pyplot as plt
V, E = [], []
for atoms in read('Pt.traj@:'):
V.append(atoms.get_volume() / len(atoms))
E.append(atoms.get_potential_energy() / len(atoms))
eos = EOS(V, E, 'birchmurnaghan')
eos.fit(warn=False)
fig = plt.figure()
ax = fig.gca()
eos.plot(ax=ax) # draw onto the current axes
ax.set_xlim(14.0, 16.0)
ax.set_xlabel('volume [Å^3/atom]')
ax.set_ylabel('energy [eV/atom]')
plt.tight_layout()
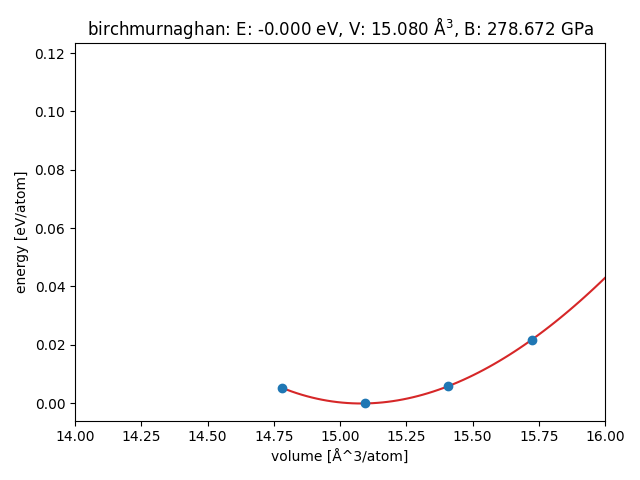
Result for Pt using EMT compared to experiment and WIEN2k Equilibrium volumes (Å^3/atom):
symbol |
emt |
exp |
wien2k |
|---|---|---|---|
Pt |
15.08 |
15.02 |
15.64 |
Al |
15.93 |
16.27 |
16.48 |
Ni |
10.60 |
10.81 |
10.89 |
Au |
16.68 |
16.82 |
17.97 |
Pd |
14.59 |
14.56 |
15.31 |
Ag |
16.77 |
16.85 |
17.85 |
Cu |
11.57 |
11.65 |
11.95 |
Bulk moduli in GPa:
Pt |
278.67 |
285.51 |
248.71 |
Al |
39.70 |
77.14 |
78.08 |
Ni |
176.23 |
192.46 |
200.37 |
Au |
174.12 |
182.01 |
139.11 |
Pd |
180.43 |
187.19 |
168.63 |
Ag |
100.06 |
105.71 |
90.15 |
Cu |
134.41 |
144.28 |
141.33 |
Pressure derivative of bulk-moduli:
Pt |
5.31 |
5.18 |
5.46 |
Al |
2.72 |
4.45 |
4.57 |
Ni |
3.76 |
4.00 |
5.00 |
Au |
5.46 |
6.40 |
5.76 |
Pd |
5.17 |
5.00 |
5.56 |
Ag |
4.75 |
4.72 |
5.42 |
Cu |
4.21 |
4.88 |
4.86 |
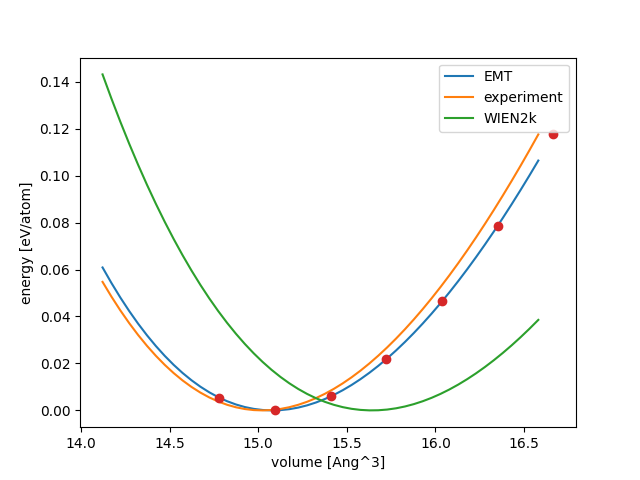
Now, we can calculate \(\Delta\) between EMT and WIEN2k for Pt:
np.float64(0.03205389052984125)
Here are all the \(\Delta\) values (in meV/atom) calculated with the script below:
Pt |
3.5 |
32.2 |
35.9 |
Al |
5.9 |
8.6 |
3.6 |
Ni |
8.6 |
12.5 |
3.7 |
Au |
5.9 |
43.7 |
39.4 |
Pd |
1.0 |
27.6 |
29.0 |
Ag |
1.9 |
22.4 |
21.3 |
Cu |
2.7 |
11.9 |
9.5 |
from ase.eos import birchmurnaghan
# Read EMT data:
data = json.loads(Path('fit.json').read_text())
# Insert values from experiment and WIEN2k:
for symbol in data:
dcdft_dct = dcdft.data[symbol]
dcdft_dct['exp_B'] *= 1e-24 * kJ
dcdft_dct['wien2k_B'] *= 1e-24 * kJ
data[symbol].update(dcdft_dct)
for name in ['volume', 'B', 'Bp']:
with open(name + '.csv', 'w') as fd:
print('# symbol, emt, exp, wien2k', file=fd)
for symbol, dct in data.items():
values = [
dct[code + '_' + name] for code in ['emt', 'exp', 'wien2k']
]
if name == 'B':
values = [val * 1e24 / kJ for val in values]
print(
f'{symbol},',
', '.join(f'{value:.2f}' for value in values),
file=fd,
)
with open('delta.csv', 'w') as fd:
print('# symbol, emt-exp, emt-wien2k, exp-wien2k', file=fd)
for symbol, dct in data.items():
# Get v0, B, Bp:
emt, exp, wien2k = (
(dct[code + '_volume'], dct[code + '_B'], dct[code + '_Bp'])
for code in ['emt', 'exp', 'wien2k']
)
print(
f'{symbol}, {delta(*emt, *exp) * 1000:.1f}, '
f'{delta(*emt, *wien2k) * 1000:.1f}, '
f'{delta(*exp, *wien2k) * 1000:.1f}',
file=fd,
)
if symbol == 'Pt':
va = min(emt[0], exp[0], wien2k[0])
vb = max(emt[0], exp[0], wien2k[0])
v = np.linspace(0.94 * va, 1.06 * vb)
for (v0, B, Bp), code in [
(emt, 'EMT'),
(exp, 'experiment'),
(wien2k, 'WIEN2k'),
]:
plt.plot(v, birchmurnaghan(v, 0.0, B, Bp, v0), label=code)
e0 = dct['emt_energy']
V = []
E = []
for atoms in read('Pt.traj@:'):
V.append(atoms.get_volume() / len(atoms))
E.append(atoms.get_potential_energy() / len(atoms) - e0)
plt.plot(V, E, 'o')
plt.legend()
plt.xlabel('volume [Ang^3]')
plt.ylabel('energy [eV/atom]')
plt.savefig('Pt.png')